SITE MAP
-
Provider
-
Patient
-
How to order
-
Contact us
provider
01
About the test
Other prenatal tests are available after mid pregnant and its accurate rate is lower. Also, diagnostic tests such
as
amniocentesis and chorionic villus sampling are invasive method and have the risk of miscarriage. For this, many
feel afraid.
On the other hand, NICE® is safe using maternal blood without the risk of miscarriage or rupture of membrane
caused
by invasive prenatal test and accurate with over 99% detection rate.

-

-
01-2
What is NICE®
prenatal test?NICEⓇ is a non-invasive prenatal test (NIPT) that detects fetal DNA in maternal plasma during pregnancy through Next Generation Sequencing (NGS). It can be tested from the 10th week of pregnancy and evaluates fetal chromosomal abnormalities. NICEⓇ screens for common trisomy (such as 21, 18, 13), sex chromosome aneuploidies and analyzes eight clinically important microdeletion regions.
-
02
How it works
how is the NICE® performed
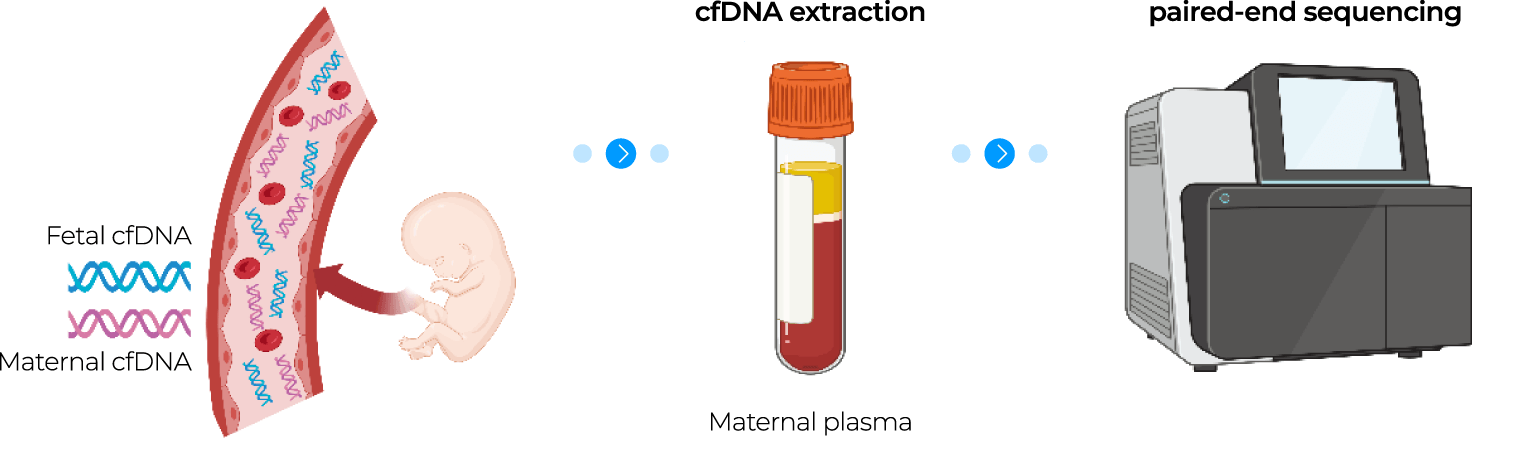
NICE® looks at isolated fetal cfDNA present in maternal blood, collected after 10 weeks of pregnancy from massively parallel WGS. Sequencing data is analyzed by applying the bioinformatics pipeline.
-
03
Test
options -
- nice® lite
- nice® basic
- nice® premium
- T13, T18, T21



- T9, T16, T22


- All chromosome

- *8 Microdeletions
- *116 Microdeletions
- *Sex Chromosome Disorder
*Any or all can be added to LITE, BASIC or PREMIUM service
04
How is the NICE® performed
NICE® looks at isolated fetal cfDNA present in maternal blood, collected after 10 weeks of pregnancy from
massively parallel WGS. Sequencing data is analyzed by applying the bioinformatics pipeline.
After removing the GC bias, normalized reads are used for further analysis. The size selection method is used to
separate and analyze fetal-derived cfDNA and maternal-derived cfDNA according to the size distribution of cfDNA.
To detect chromosomal aneuploidies, we use a multi-Z method with 21 z-scores for each autosomal chromosome. First,
fetal sex is determined using Y-derived reads, and the fetal fraction is analyzed using sex-specific
bioinformatics pipelines.
04-1 Pre-processing (Sequencing)
Figure 1. outlines the bioinformatics process from maternal blood collection to reporting of test results.
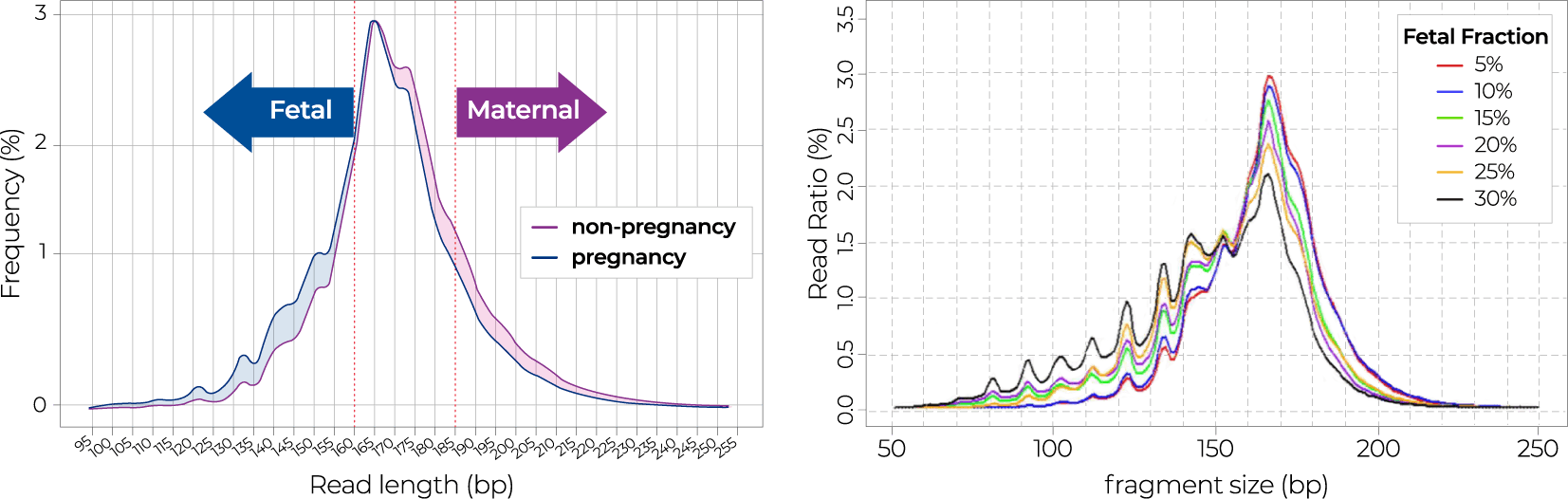
04-2 Size selection method
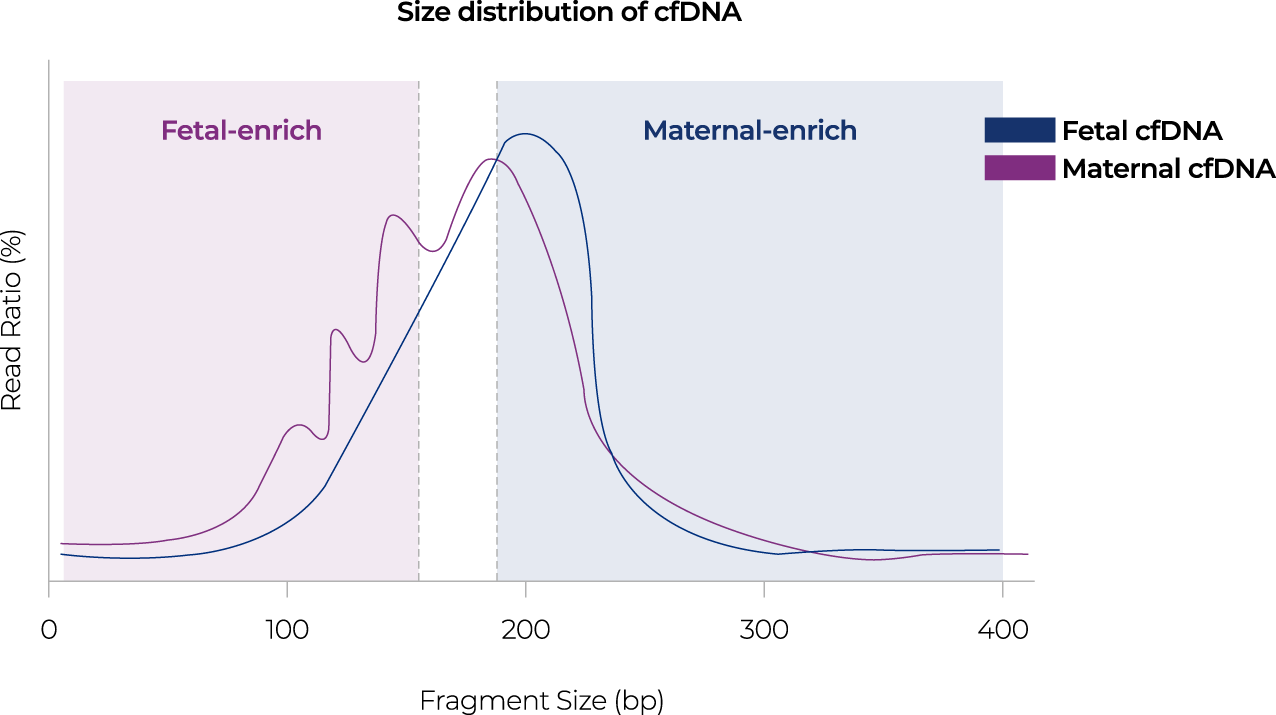
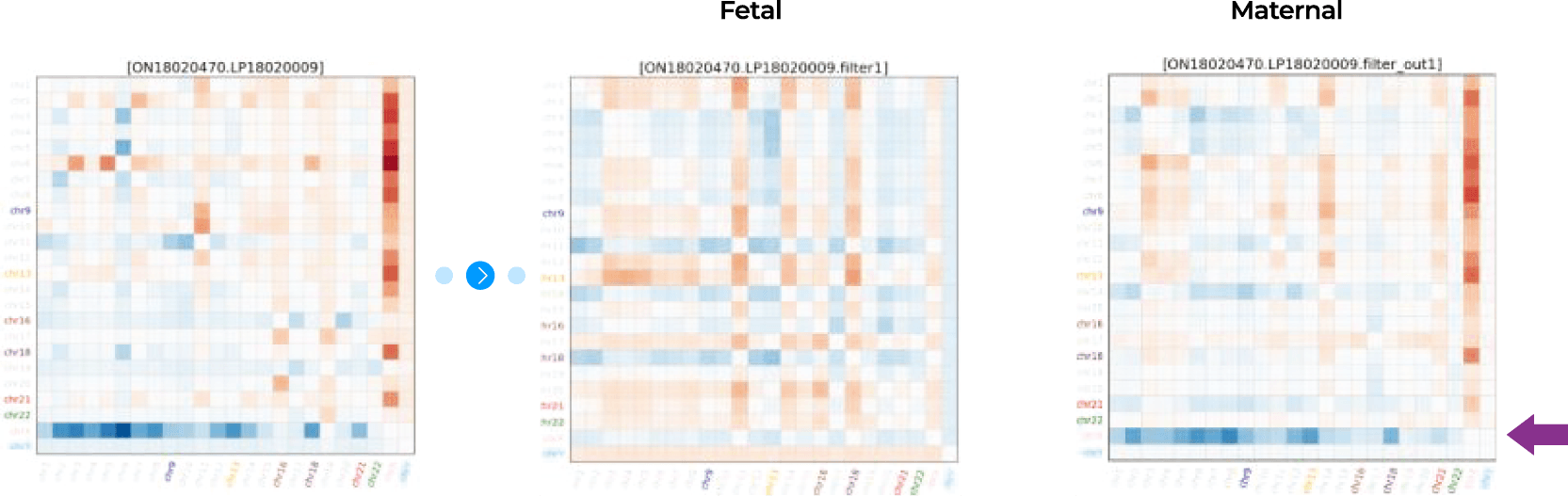
04-3 Bioinformatics anslysis
Figure 2. The workflow of Nice® pipeline based on size selection method using cfDNA fragment size.

05
Why NICE® is different
If there is an abnormality in the chromosomes derived from the placenta or the mother, false negatives and false positives may appear as a result of NIPT. Although the proportion of false- negative and false-positive results in NIPT are very small, the reporting of false results cannot be ignored in clinical setting.
Table1. Differences between NICE® and targeted sequencing methods
using 21 z-score thresholds
differences between ethnicities
cfDNA by size selection method using paired-end sequencing
maternal-derived cfDNA
Multi-Z method
-
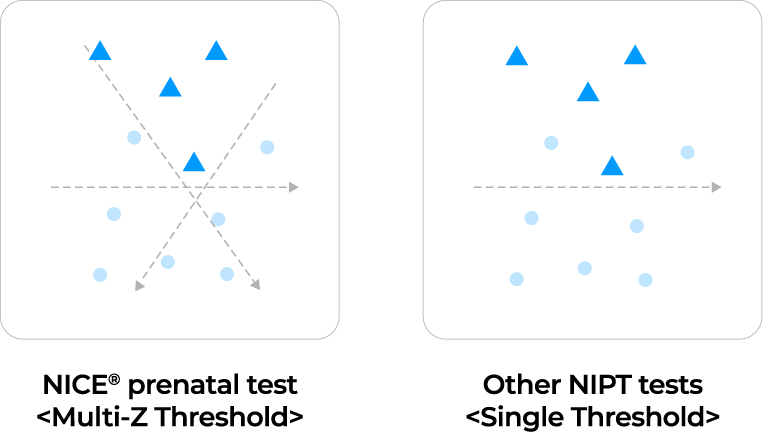
The multi-Z method has the advantage of being more precise by using a 21 z-scores compared to the existing methods using one or two z-score thresholds by other labs.
-
Size selection method
The bioinformatics pipeline of the NICE test aimed to improve the accuracy of NIPT by dividing and analyzing fetal or maternal cfDNA based on cfDNA size differences. Through the size selection method, fetal-derived cfDNA analysis had the effect of increasing the fetal DNA fraction, and maternal-derived cfDNA analysis had the effect of removing factors affecting maternal chromosomal abnormalities and increased NIPT accuracy. In addition, when gDNA is contaminated, the distribution of cfDNA fragments is different, so the quality of sequencing data is controlled through the distribution of cfDNA.
-

01 XO Case : Not detected (Maternal Mosaicism)
02 Trisomy 18 : 2-step confirmation




